The realities of working in a forest: Panama resampling of Macrosystem tree plots underway . . .
Posted: March 20, 2013 Filed under: Uncategorized Leave a commentOur lab field crew is hard at work in the heat of Barro Colorado Island recensusing our NSF Macrosystems tree plot. These data will be used to assess latitudinal variation in tree growth demography and forest turnover. Here is a photo of a very large treefall in one of our plots that (unfortunately) didn’t kill many stems… just made them much harder to find! Here, Colby and Ben are both balanced several feet off the ground in search for smaller saplings underneath all of this mess.
First day of spring . . . . help collect data and phenologize!
Posted: March 20, 2013 Filed under: Uncategorized Leave a commentHonorary lab member Carolyn Enquist (ok a little nepotism at work here!) is happy to announce that The National Phenology Network has just released a handy web tool and new fun iPhone App that let’s you collect, enter, view phenology data. Its so easy to sign up and start recording data. Check it out by clicking here. We are monitoring several trees at our house and you can search for our data collection – (our backyard Prosopis just burst its leaves yesterday !). I’m impressed by the web interface with their database, data visulalization tools, as well as the ability to search for records. NPN’s efforts are important especially seeing that in an age of budget cutbacks for basic observational research (that will likely have deep impacts on
the progress of Science). Citizen Science is likely going to be (well it increasingly is) a foundational tool for data collection and for doing Science. NPN I think may have set the bar for how to design and implement impressive cyberinfrastructure and create a fun tool to enter basic natural history observations and explore natural history via phenology. So in honor of the vernal equinox, sign up and use Nature’s Notebook! Help them reach there goal of 1 million observations this year.
Sign up if you have not already to the “Introduction to programing for Biologists” boot camp
Posted: March 19, 2013 Filed under: Uncategorized 1 CommentThis will undoubtably be a useful short course. A big congratulations to lab member Julie Messier for bringing this short course to campus. Here is the link to the registration page for the Intro to programming for biologists workshop. It will be offered by the software carpentry group on the UA Campus on April 4th and 5th. http://www.eventbrite.com/event/5396735782
The course will give priority to people working with organismal biology/ecology until TODAY (Tuesday March 18th), then we will open up registration more broadly to everybody.
There is a 20$ fee associated with the workshop to ensure that people that sign up do indeed attend both days of the workshop.
Thanks again Julie for spearheading this effort!
Global Temperatures Highest in 4,000 Years
Posted: March 8, 2013 Filed under: Uncategorized 1 CommentFascinating new study in Science showing the magnitude of the influence of human released greenhouse gases on Earth’s climate. This part of the study caught my eye . . .
“Scientists say that if natural factors were still governing the climate, the Northern Hemisphere would probably be destined to freeze over again in several thousand years. “We were on
this downward slope, presumably going back toward anoth
er ice age,” Dr. Marcott said. Instead, scientists believe the enormous increase in greenhouse gases caused by industrialization will almost certainly prevent that.” In other words, the current increase in greenhouse gases is enough to reverse the ‘natural’ oscillations of climate. Incredible.
Simulation versus analytical approaches? The forgotten merits of the analytic viewpont.
Posted: March 5, 2013 Filed under: Uncategorized 3 CommentsPostdoc Lisa Patrick Bentley forwarded a fun paper to me this week. The debate about how to best approach complex problems in ecology and evolution is an old one. It seems that one can often find oneself in a debate over the merits of a mathematical model verus a simulation model. In the lab we have used both approaches.
In latest issue of Eos there is a nice piece on “The Forgotten Merits of the Analytical Viewpoint” by K.C. Lewis. Lewis reminds us that although computer simulation is easy and powerfully insightful the analytical approach yields unique insights.
She states “In this age of powerful computers, researchers use massively parallelized codes that include nonlinear effects and multiple layers of strongly coupled physicalprocesses. With such cap ability, many see the analytic approach as outmoded or, at best, as a way to validate codes in simple scenarios. It seems that many of the strengths of analytic methods have been largely forgotten.” and concludes that “. . .just as experimental results should be interpreted in the light of theory, so should results derived from simulations. If nature itself is seen as a sophisticated numerical simulation where every possible effect has been included, the past successes of analytical models in the study of nature argue that attempts to match numerical simulation results to analytic solutions will often be successful. Due to the theoretical advantages mentioned above for the analytic viewpoint, this success will often mean increased physical insight.”

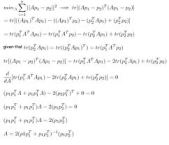
*** a careful reader pointed out that the author of this paper is a she and not a he! My sincere apologies to the author and readers for not catching this the first time.
Keeping up with the literature . . .
Posted: January 31, 2013 Filed under: Uncategorized 3 Comments Perhaps the most important but daunting task with starting graduate school (or a well seasoned faculty member!) is keeping up with the literature. Today we had a quick lab tutorial at lab meeting to how to best optimize access to the literature as well as how to keep on top of the deluge of papers being published. Not many people know about these tips even though google scholar has been around for a while. Don’t take my word from it but read it from Jon Eisen. Here is a quick summary of some of the areas covered . .
Perhaps the most important but daunting task with starting graduate school (or a well seasoned faculty member!) is keeping up with the literature. Today we had a quick lab tutorial at lab meeting to how to best optimize access to the literature as well as how to keep on top of the deluge of papers being published. Not many people know about these tips even though google scholar has been around for a while. Don’t take my word from it but read it from Jon Eisen. Here is a quick summary of some of the areas covered . .
(1) Stop entering your references by hand. . . Go to google scholar. Click on settings in the upper left hand column. At the bottom of the first page, click on “show links to import citations into. . . ” Easier to upload and manage citations. You will notice also that there is a ‘More’ link at the end of the listing for a specific paper in google scholar. Click on it to get a preformated reference that you can cut and paste if needed. Or click on the ‘Import into BibTeX’ or ‘Important into EndNote’ link. Also, if you search for a journal article most journal websites will have a link to download the reference into the reference manager you likely use.
(2) Make sure you can access journal articles anywhere! On or off campus make google work for you. On the same page as above, also visit the upper left hand side, and click on “Library links”. Here search for your instutution library. For us we put in ‘University of Arizona’ but you can choose other libraries too. Make sure to then click on your library when it is found but also click ‘save’ at the bottom of the page. Now, when you search for a paper in google scholar you will get a link to that paper to the library that you specify. When you click on that library, in the case of our UofA library, we automatically login with our netid which then allows us access papers anywhere off campus or even overseas.
(3) Make a scholar profile. If you have not done so, create a google scholar profile. You don’t have to make it a public profile but I would encourage you to do s0 as others can more actively follow your work. See the official google instructions here. Also see here or here for help.
(4) Want to get an automated summary of new papers published by authors you are really interested in? It is easy in google scholar – find a researcher profile you are interested in (granted they need to have set up their profile). For example see David Tilman‘s link. Interested in being alterted when ever he publishes a paper? even in a journal you don’t regularly browse? Then click the “Follow new articles” link on the right hand side and fill in the information. If you are really into Tilman then click on ‘Follow new citations’ and google will send you a sumarized digest of new papers who are citing Tilman (that will likely be a long email!). You can manage your ‘citation alerts’ in your google scholar account main page in the upper right hand corner.
(5) Want to know everytime who is citing your favorite paper in the world? In google scholar search a paper that you want to stay on top of who is citing that paper (all papers that are published that cite the paper you are interested in). You will see a link on the google scholar search results page for that paper in the lower left hand side ‘Create alert’. Click on it and fill on the information. Now you will get a email digest every week or so (or less if few people cite the paper) from google letting you know about papers that are recently published that are citing that paper. This is a good way to stay on top of a field that revolves around key papers.
(6) Let the google universe bring the literature you should be reading to you . . . Not sure how they do this but once you set up your google scholar account then check out the google scholar front page, click on the bell icon in the upper right. You will be directed to a new page – based on your citations (not sure if you need to have several citations of your own publications for this to work) – full of papers that google is recommending. Their recommendations of papers for you looks to be based on your publications and who is citing them. I’ve discovered many, many papers here that I would not have normally run into. Neat stuff. Google states officially here
“We analyze your articles (as identified in your Scholar profile), scan the entire web looking for new articles relevant to your research, and then show you the most relevant articles when you visit Scholar. We determine relevance using a statistical model that incorporates what your work is about, the citation graph between articles, the fact that interests can change over time, and the authors you work with and cite. You don’t need to configure updates or enter any queries. We’ll notify you about new updates by displaying a preview on the homepage and highlighting a bell icon on search results pages:”
(7) Search efficiently . . . I’m surprised by how many students do not know how to make the most out of google scholar. See here or even here to learn how to be more efficient at searching. Here are some other links that may be of some help. Morgan Ernest also had a nice post here.
Hot, rapid fire talks at ESA this year . . . .
Posted: January 31, 2013 Filed under: Uncategorized Leave a comment I’m encouraged that the Ecological Society of America is continuing to push and inject new enthusism into the annual meeting. This year at the ESA meeting look out for the new Ignite ESA Sessions. Here is a description from the ESA website, which sounds crazy (in a good way) . . .
I’m encouraged that the Ecological Society of America is continuing to push and inject new enthusism into the annual meeting. This year at the ESA meeting look out for the new Ignite ESA Sessions. Here is a description from the ESA website, which sounds crazy (in a good way) . . .
“Ignite ESA Sessions will be scheduled for 1 hour and consist of up to 10 5-minute talks presented in the Ignite style (http://igniteshow.com/) which address a common theme. Each Ignite talk will feature 20 slides that advance automatically every 15 seconds. Although there will be no time during the session for questions and answers, there will be a designated space nearby where presenters and attendees will be encouraged to gather for extended discussion immediately after each session. Each session should be organized around a general theme that is related to the advancement of ecology, either conceptually or technologically“
Charles Darwin wished he had worked harder at mathematics
Posted: January 25, 2013 Filed under: Uncategorized Leave a comment What is it like to do fun Science? Here is a compelling documentary produced at the Santa Fe Institute – covers some good biology, ecology, history, the role of math, art, and modeling in science. “Charles Darwin wrote that he wished he had worked harder at mathematics, so as to possess the “extra sense” that he felt mathematicians can bring to understanding the world. The new 45 minute documentary, “Darwin’s Extra Sense”, explores how twenty-first century mathematics is bringing an extra sense to the study of the life sciences.”
What is it like to do fun Science? Here is a compelling documentary produced at the Santa Fe Institute – covers some good biology, ecology, history, the role of math, art, and modeling in science. “Charles Darwin wrote that he wished he had worked harder at mathematics, so as to possess the “extra sense” that he felt mathematicians can bring to understanding the world. The new 45 minute documentary, “Darwin’s Extra Sense”, explores how twenty-first century mathematics is bringing an extra sense to the study of the life sciences.”
Why you should use this tool to standardize your species names …
Posted: January 24, 2013 Filed under: Uncategorized Leave a commentNew lab publication – the TNRS! The Taxonomic Name Resolution Service is now officially published. Check out the paper now in BMC Bioinformatics. 
BACKGROUND: The digitization of biodiversity data is leading to the widespread application of taxon names that are superfluous, ambiguous or incorrect, resulting in mismatched records and inflated species numbers. The ultimate consequences of misspelled names and bad taxonomy are erroneous scientific conclusions and faulty policy decisions. The lack of tools for correcting this ‘names problem’ has become a fundamental obstacle to integrating disparate data sources and advancing the progress of biodiversity science.
RESULTS: The TNRS, or Taxonomic Name Resolution Service, is an online application for automated and user-supervised standardization of plant scientific names. The TNRS builds upon and extends existing open-source applications for name parsing and fuzzy matching. Names are standardized against multiple reference taxonomies, including the Missouri Botanical Garden’s Tropicos database. Capable of processing thousands of names in a single operation, the TNRS parses and corrects misspelled names and authorities, standardizes variant spellings, and converts nomenclatural synonyms to accepted names. Family names can be included to increase match accuracy and resolve many types of homonyms. Partial matching of higher taxa combined with extraction of annotations, accession numbers and morphospecies allows the TNRS to standardize taxonomy across a broad range of active and legacy datasets.
CONCLUSIONS: We show how the TNRS can resolve many forms of taxonomic semantic heterogeneity, correct spelling errors and eliminate spurious names. As a result, the TNRS can aid the integration of disparate biological datasets. Although the TNRS was developed to aid in standardizing plant names, its underlying algorithms and design can be extended to all organisms and nomenclatural codes. The TNRS is accessible via a web interface at http://tnrs.iplantcollaborative.org/ and as a RESTful web service and application programming interface. Source code is available at https://github.com/iPlantCollaborativeOpenSource/TNRS/.
New lab post doc!
Posted: August 17, 2012 Filed under: Uncategorized Leave a comment
A big warm welcome to Sean Michaletz who is joining our lab this month! He is one of the post docs associated with the Macrosytems project. Here Sean (in the red hat) is being introduced to the wonders of the Gentry plot, tree paint, and contemplating how to bring down leaves and stem traits from a rather large trees with branches out of seeming reach of any human. . . .






